OraMod First technical Review
The OraMod Consortium will present the results of the first year of research to the European Commission in Brussels.
Read more →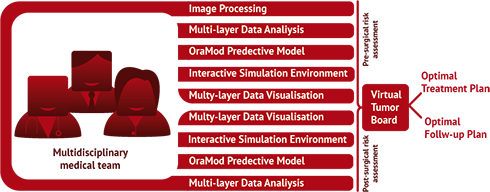
OraMod supports clinicians in personalized management of patients. The multidisciplinary medical team (Tumor Board) can virtually interact to plan personalized treatment and follow-up pathways based on a shared view of all patients' data.
OraMod is a European Project co-funded under the European Community’s 7th Framework Programme devoted to support the clinical specialists in the management of patients diagnosed with Oral Cavity Cancer, to improve survival and reduce impacts on patients' life.
The projects focuses on the early prediction of patients at high risk for oral cavity cancer reoccurrence, so enabling personalized disease management. OraMod innovative methods, tools, models and predictive markers will help clinicians to manage and analyze the big amount of information related to disease manifestation in each patient and will support their treatment decisions.
In so doing the project will facilitate the multi-specialist approach to diagnosis, risk assessment, and treatment decisions and will realize the integration of research-derived evidences into the clinical practice (i.e. the evidence-driven approach).
The OraMod Consortium will present the results of the first year of research to the European Commission in Brussels.
Read more →OraMod Project was presented at the EACMFS Congress of European Association for Cranio Maxillo-Facial Surgery in Prague, Czech Republic from 23rd to 26th September 2014.
Read more →OraMod Project was presented at the SPIE Medical Imaging Conference which was held in San Diego, California (USA) from 15th to 20th February 2014.
Read more →
OraMod has been presented to the public by the coordinator University of Parma. A press release is published on the local Gazzetta di Parma newspaper and on the university web site.
Read more →The OraMod Consortium will be meeting in Amsterdam on February 13th-14th, 2014 at VU medical center. The meeting will consolidate the clinical study and the OraMod predictive model and VPH platform.
Read more →The OraMod Consortium in October 2014 will hold its 4th meeting at Heinrich-Heine University Dusseldorf.
Read more →GET is a Collaborative Support Action co-funded by the European Commission, which aims at maximizing the impact of project results.
Read more →The second BHI International Conference on Biomedical and Health Informatics will be held in Valencia (Spain) from the 1st to the 4th of June 2014.
Read more →OraMod clinicians will participate to the Oral Cancer Day 2014 to inform citizens regarding oral cancer risks and to promote the oral cancer prevention campaign which will involve more than 8.000 dentists and surgeons in Italy only.
Read more (Italian) →